Difference between revisions of "How to add transport of N solutes to icoFoam"
(→Files) |
AntonChupin (Talk | contribs) |
||
| (7 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
Before reading this tutorial, you should first understand [[How to add temperature to icoFoam | how to add temperature to icoFoam]]. The idea here is the analogous, but in the case of [http://en.wikipedia.org/wiki/Solution solutions] one can have an indefinite number of '''solutes''' dissolved in a '''[http://en.wikipedia.org/wiki/Solvent solvent]'''. The dissolution of different phases is not modelled here, therefore solute concentrations may not increase above [http://en.wikipedia.org/wiki/Solubility solubility saturation] (under saturation hypothesis). | Before reading this tutorial, you should first understand [[How to add temperature to icoFoam | how to add temperature to icoFoam]]. The idea here is the analogous, but in the case of [http://en.wikipedia.org/wiki/Solution solutions] one can have an indefinite number of '''solutes''' dissolved in a '''[http://en.wikipedia.org/wiki/Solvent solvent]'''. The dissolution of different phases is not modelled here, therefore solute concentrations may not increase above [http://en.wikipedia.org/wiki/Solubility solubility saturation] (under saturation hypothesis). | ||
| − | == | + | == solutionProperties dictionary == |
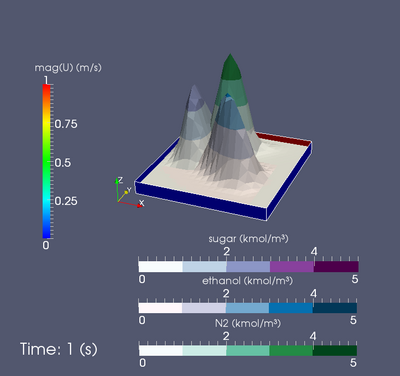
| + | [[Image:CavityTransportIcoFoamN.png|400px|thumb|right|Cavity example of transportIcoFoamN solver.]] | ||
The solute list containing [http://en.wikipedia.org/wiki/Fick's_laws_of_diffusion Fick's Law] diffusion coefficients is defined in | The solute list containing [http://en.wikipedia.org/wiki/Fick's_laws_of_diffusion Fick's Law] diffusion coefficients is defined in | ||
case/constant/solutionProperties | case/constant/solutionProperties | ||
| Line 24: | Line 25: | ||
} | } | ||
</cpp> | </cpp> | ||
| + | (Obs.: diffusionModel is not used here, but as an exercise you could model a temperature sensitive diffusion or a diffusivity model for turbulent flow, etc.) | ||
| − | + | == Time mesh dictionaries == | |
| + | You will have to create field dictionaries for each solute (e.g. sugar, ethanol, etc.). Because each solute field has dimensions of molar concentration (kmol/m³), the field names are defined as "C_{solute}" (e.g. 0/C_sugar). | ||
| + | |||
| + | == setFields == | ||
| + | I used ''setFields'' utility to set different concentrations in the internal field. Check | ||
| + | cavity/system/setFieldsDict | ||
| + | |||
| + | == Container class ''solute'' == | ||
| + | Now it is a good opportunity learn a little bit of class inheritance. Let's create a container class for each solute, where the diffusivity constants (D) are stored together with the ''volScalarFields'' (C_solute). The ''solute'' class inherits all properties of ''volScalarFields'' -- and was based on the "phase" class used in multiphase solvers. | ||
| + | |||
| + | The inheritance is defined in ''solute.H'' by declaring | ||
| + | <cpp> | ||
| + | class solute | ||
| + | : | ||
| + | public volScalarField | ||
| + | </cpp> | ||
| + | |||
| + | The ''solute'' is constructed entering the solute dictionary (sub-dictionary in solutionProperties-file) and the mesh. | ||
| + | <cpp> | ||
| + | Foam::solute::solute | ||
| + | ( | ||
| + | const dictionary& soluteDict, | ||
| + | const fvMesh& mesh | ||
| + | ) | ||
| + | : | ||
| + | volScalarField | ||
| + | ( | ||
| + | IOobject | ||
| + | ( | ||
| + | "C_" + soluteDict.dictName(), | ||
| + | mesh.time().timeName(), | ||
| + | mesh, | ||
| + | IOobject::MUST_READ, | ||
| + | IOobject::AUTO_WRITE | ||
| + | ), | ||
| + | mesh | ||
| + | ), | ||
| + | name_("C_" + soluteDict.dictName()), | ||
| + | soluteDict_(soluteDict), | ||
| + | D_(soluteDict_.lookup("D")), | ||
| + | diffusionModel_(soluteDict_.lookup("diffusionModel")) | ||
| + | { | ||
| + | Info<< "Reading field " << name_ <<"\n" << endl; | ||
| + | } | ||
| + | </cpp> | ||
| + | |||
| + | We are going to dynamically allocate memory to each solute. Therefore we will be using pointers, which makes it interesting to have an instances ''solute::clone()'' and ''solute::iNew''. | ||
| + | |||
| + | Finally, don't forget to declare ''solute'' in the main solver header. | ||
| + | <cpp> | ||
| + | #include "solute.H" | ||
| + | </cpp> | ||
| + | |||
| + | == createFields.H == | ||
| + | First, let's have a look into ''createFields.H'' to understand how ''solutionProperties'' dictionary and the C_-fields are read. | ||
| + | <cpp> | ||
| + | // * * * * * * * * * * * * * Properties fields * * * * * * * * * * * * * * * // | ||
| + | |||
| + | Info<< "Reading solutionProperties\n" << endl; | ||
| + | |||
| + | IOdictionary solutionProperties | ||
| + | ( | ||
| + | IOobject | ||
| + | ( | ||
| + | "solutionProperties", | ||
| + | runTime.constant(), | ||
| + | mesh, | ||
| + | IOobject::MUST_READ_IF_MODIFIED, | ||
| + | IOobject::NO_WRITE | ||
| + | ) | ||
| + | ); | ||
| + | |||
| + | const wordList soluteNameList(solutionProperties.toc()); | ||
| + | |||
| + | PtrList<solute> soluteList(soluteNameList.size()); | ||
| + | |||
| + | forAll(soluteNameList, i) | ||
| + | { | ||
| + | const word& soluteName = soluteNameList[i]; | ||
| + | const dictionary& soluteDict = solutionProperties.subDict(soluteName); | ||
| + | |||
| + | soluteList.set | ||
| + | ( | ||
| + | i, | ||
| + | new solute | ||
| + | ( | ||
| + | soluteDict, | ||
| + | mesh | ||
| + | ) | ||
| + | ); | ||
| + | } | ||
| + | </cpp> | ||
| + | |||
| + | ''IOdictionary::toc()'' returns a wordList containing the table of contents (toc) of ''solutionProperties''. | ||
| + | |||
| + | Next, a list of pointers to solute class having the size of the number of entries in the ''solutionProperties'' dictionary is defined, before dynamically allocating solutes in the list by the "new" operator. | ||
| + | |||
| + | == Groups for discretization schemes and solution options == | ||
| + | We want to play with an indefinite number of N solutes, so it would be very troublesome to handle the solution options of those N solutes in ''fvSolution'' and ''fvSchemes'' dictionaries. To avoid that we can define the solver group | ||
| + | <cpp> | ||
| + | C_solutes | ||
| + | { | ||
| + | solver BICCG; | ||
| + | preconditioner DILU; | ||
| + | tolerance 1e-7; | ||
| + | relTol 0; | ||
| + | }; | ||
| + | </cpp> | ||
| + | |||
| + | and the divergent and laplacian schemes: | ||
| + | <cpp> | ||
| + | div(phi,C_solutes) Gauss upwind; | ||
| + | </cpp> | ||
| + | and | ||
| + | <cpp> | ||
| + | laplacian(D,C_solutes) Gauss linear corrected; | ||
| + | </cpp> | ||
| + | |||
| + | The group's dictionaries can be called by the ''fvm::div'' and ''fvm::laplacian'' and the solver dictionary must be given to ''fvScalarMatrix::solver'' (see below). | ||
| + | |||
| + | <cpp> | ||
| + | // --- Mass transport | ||
| + | |||
| + | dictionary soluteSolveDict = mesh.solutionDict().subDict("solvers").subDict("C_solutes"); | ||
| + | forAll(soluteNameList, i) | ||
| + | { | ||
| + | fvScalarMatrix MassTransport | ||
| + | ( | ||
| + | fvm::ddt(soluteList[i]) | ||
| + | + fvm::div(phi, soluteList[i], "div(phi,C_solutes)") | ||
| + | - fvm::laplacian(soluteList[i].D(), soluteList[i], "laplacian(D,C_solutes)") | ||
| + | ); | ||
| + | |||
| + | MassTransport.solve(soluteSolveDict); | ||
| + | } | ||
| + | </cpp> | ||
| + | |||
| + | === Using POSIX regular expressions === | ||
| + | You may also find it useful that OpenFOAM uses [http://en.wikipedia.org/wiki/Regular_expression regular expressions], for example you can specify: | ||
| + | <cpp> | ||
| + | "C_solute1|C_solute2" | ||
| + | { | ||
| + | solver BICCG; | ||
| + | preconditioner DILU; | ||
| + | tolerance 1e-7; | ||
| + | relTol 0; | ||
| + | }; | ||
| + | </cpp> | ||
== Files == | == Files == | ||
Source files and example case: [[File:TransportIcoFoamN.tar.gz]] | Source files and example case: [[File:TransportIcoFoamN.tar.gz]] | ||
| + | |||
| + | == References == | ||
| + | *[http://www.cfd-online.com/Forums/openfoam-programming-development/85049-read-properties-per-component-input-file-dictionary.html CFD Online forum: Read properties per component from an input file dictionary] | ||
| + | *[http://www.cfd-online.com/Forums/openfoam-programming-development/105864-lookup-non-included-scalars-list.html CFD Online forum: lookup non included scalars from a list] | ||
| + | |||
| + | [[Category:Tutorials]] | ||
Latest revision as of 11:18, 21 October 2013
Before reading this tutorial, you should first understand how to add temperature to icoFoam. The idea here is the analogous, but in the case of solutions one can have an indefinite number of solutes dissolved in a solvent. The dissolution of different phases is not modelled here, therefore solute concentrations may not increase above solubility saturation (under saturation hypothesis).
Contents
1 solutionProperties dictionary
The solute list containing Fick's Law diffusion coefficients is defined in
case/constant/solutionProperties
Check the example below
sugar { diffusionModel constant; D D [0 2 -1 0 0 0 0] 5.2e-10; } ethanol { diffusionModel constant; D D [0 2 -1 0 0 0 0] 1.6e-9; } N2 { diffusionModel constant; D D [0 2 -1 0 0 0 0] 3.6e-9; }
(Obs.: diffusionModel is not used here, but as an exercise you could model a temperature sensitive diffusion or a diffusivity model for turbulent flow, etc.)
2 Time mesh dictionaries
You will have to create field dictionaries for each solute (e.g. sugar, ethanol, etc.). Because each solute field has dimensions of molar concentration (kmol/m³), the field names are defined as "C_{solute}" (e.g. 0/C_sugar).
3 setFields
I used setFields utility to set different concentrations in the internal field. Check
cavity/system/setFieldsDict
4 Container class solute
Now it is a good opportunity learn a little bit of class inheritance. Let's create a container class for each solute, where the diffusivity constants (D) are stored together with the volScalarFields (C_solute). The solute class inherits all properties of volScalarFields -- and was based on the "phase" class used in multiphase solvers.
The inheritance is defined in solute.H by declaring
class solute
:
public volScalarField
The solute is constructed entering the solute dictionary (sub-dictionary in solutionProperties-file) and the mesh.
Foam::solute::solute ( const dictionary& soluteDict, const fvMesh& mesh ) : volScalarField ( IOobject ( "C_" + soluteDict.dictName(), mesh.time().timeName(), mesh, IOobject::MUST_READ, IOobject::AUTO_WRITE ), mesh ), name_("C_" + soluteDict.dictName()), soluteDict_(soluteDict), D_(soluteDict_.lookup("D")), diffusionModel_(soluteDict_.lookup("diffusionModel")) { Info<< "Reading field " << name_ <<"\n" << endl; }
We are going to dynamically allocate memory to each solute. Therefore we will be using pointers, which makes it interesting to have an instances solute::clone() and solute::iNew.
Finally, don't forget to declare solute in the main solver header.
#include "solute.H"
5 createFields.H
First, let's have a look into createFields.H to understand how solutionProperties dictionary and the C_-fields are read.
// * * * * * * * * * * * * * Properties fields * * * * * * * * * * * * * * * // Info<< "Reading solutionProperties\n" << endl; IOdictionary solutionProperties ( IOobject ( "solutionProperties", runTime.constant(), mesh, IOobject::MUST_READ_IF_MODIFIED, IOobject::NO_WRITE ) ); const wordList soluteNameList(solutionProperties.toc()); PtrList<solute> soluteList(soluteNameList.size()); forAll(soluteNameList, i) { const word& soluteName = soluteNameList[i]; const dictionary& soluteDict = solutionProperties.subDict(soluteName); soluteList.set ( i, new solute ( soluteDict, mesh ) ); }
IOdictionary::toc() returns a wordList containing the table of contents (toc) of solutionProperties.
Next, a list of pointers to solute class having the size of the number of entries in the solutionProperties dictionary is defined, before dynamically allocating solutes in the list by the "new" operator.
6 Groups for discretization schemes and solution options
We want to play with an indefinite number of N solutes, so it would be very troublesome to handle the solution options of those N solutes in fvSolution and fvSchemes dictionaries. To avoid that we can define the solver group
C_solutes
{
solver BICCG;
preconditioner DILU;
tolerance 1e-7;
relTol 0;
};
and the divergent and laplacian schemes:
div(phi,C_solutes) Gauss upwind;
and
laplacian(D,C_solutes) Gauss linear corrected;
The group's dictionaries can be called by the fvm::div and fvm::laplacian and the solver dictionary must be given to fvScalarMatrix::solver (see below).
// --- Mass transport dictionary soluteSolveDict = mesh.solutionDict().subDict("solvers").subDict("C_solutes"); forAll(soluteNameList, i) { fvScalarMatrix MassTransport ( fvm::ddt(soluteList[i]) + fvm::div(phi, soluteList[i], "div(phi,C_solutes)") - fvm::laplacian(soluteList[i].D(), soluteList[i], "laplacian(D,C_solutes)") ); MassTransport.solve(soluteSolveDict); }
6.1 Using POSIX regular expressions
You may also find it useful that OpenFOAM uses regular expressions, for example you can specify:
"C_solute1|C_solute2" { solver BICCG; preconditioner DILU; tolerance 1e-7; relTol 0; };
7 Files
Source files and example case: File:TransportIcoFoamN.tar.gz